GFP (Green Fluorescent Protein) – History and uses
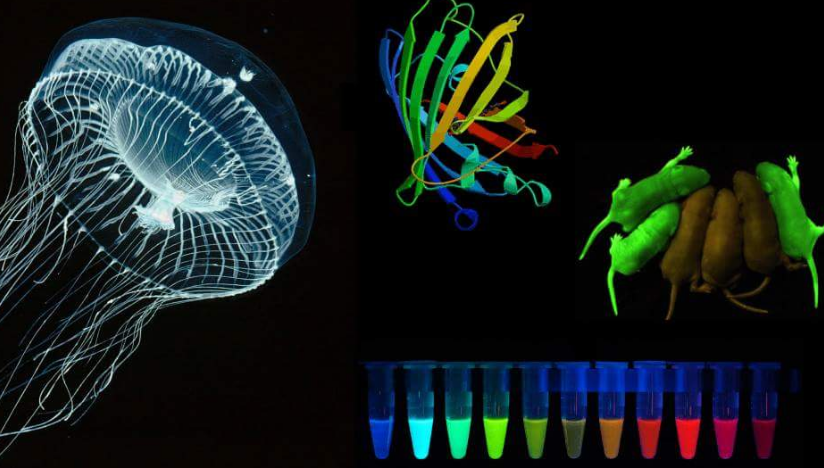
GFP is a well-known fluorescent protein, its extraction earned a Nobel Prize in Chemistry. Aequorea victoria is a jellyfish of the Hydrozoa class which can be known as a crystal jellyfish and is discovered within the seas of the West coast of North America. This species doesn’t exceed a number of centimeters in dimension and like most jellyfish is carnivorous: its tentacles have nematocysts that inject a poison able to immobilizing small prey, however innocent to people. However, the primary and most necessary attribute of this animal is its bioluminescence, i.e. it is ready to emit gentle from specific areas surrounding the hat because of chemical reactions that embrace two proteins: the aequorin protein and the GFP molecule.
Green Fluorescent Protein
Green Fluorescent Protein – gfp The GFP (in Italian fluorescent inexperienced protein), particularly, was extracted for the primary time by Osamu Shimomura, who was nominated Nobel Prize in Chemistry in 2008 for this necessary discovery. This protein, in truth, apart from having modest dimensions, if struck by radiation at a particular wavelength, re-emits a lightweight of a shiny inexperienced shade, due to this fact it may be used as a marker for the identification and subcellular localization of proteins or of specific traits within the genome and for a lot of different uses.
Needless to say, how a lot this discovery radically modified the world of fluorescence microscopy and GFP rapidly changed the far more harmful beforehand used radioactive markers; additionally, because of its comparatively easy construction, genetic engineering has managed to change it and create GFPs that emit totally different colours by fluorescence.
Recently GFP has additionally been utilized in artwork: the artist Eduardo Kac created a fluorescent inexperienced rabbit by engineering the GFP protein in its cells.
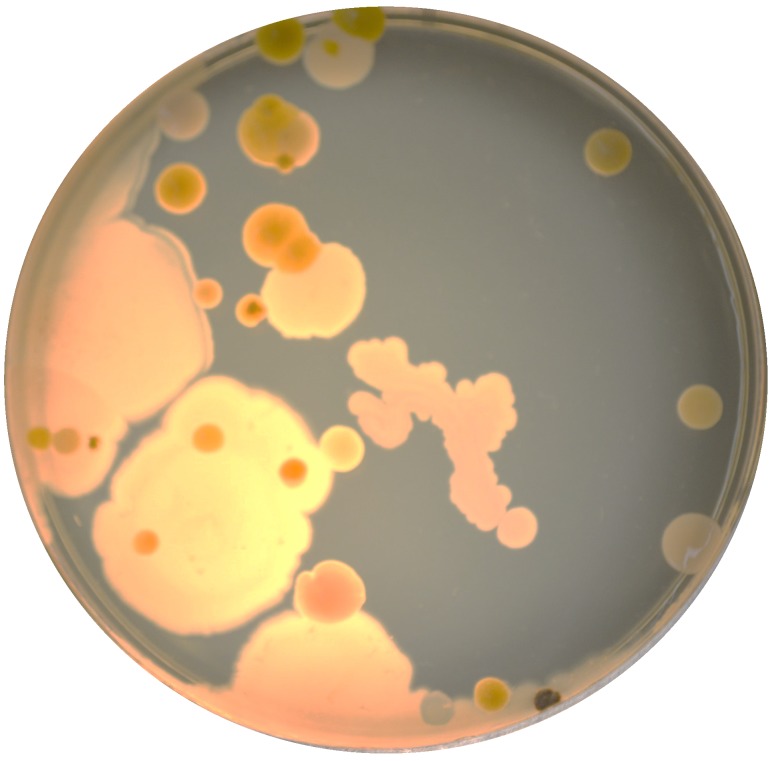
Drawing obtained on a tradition plate, crawling bacterial cultures containing totally different types of GFP (plus one other protein with crimson fluorescence).
However, engineered crops and animals are nonetheless controversial, and they’re stimulating an necessary dialogue on the protection and morality of genetic engineering.
Send suggestions