Western Blotting Protocol
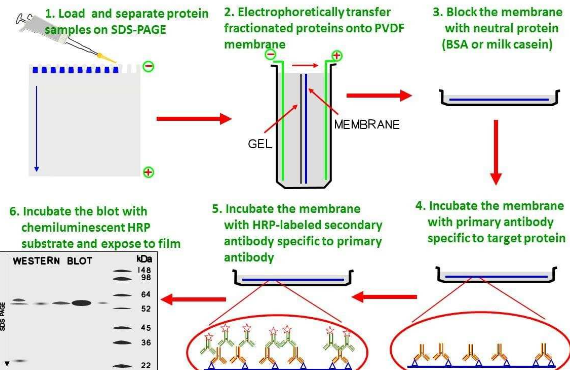
Western blotting (WB) is extensively used to research particular protein expression in cell or tissue extracts. At Cell Signaling Technology (CST) we perceive that western blotting experiments are time consuming and that their success has a essential impression in your analysis progress. For that purpose, we thoughtfully develop antibodies and supply optimized protocols together with reference data and technical assist to make your western blotting expertise profitable.
For western blots, incubate membrane with diluted major antibody in both 5% w/v BSA or nonfat dry milk, 1X TBS, 0.1% Tween® 20 at 4°C with mild shaking, in a single day.
Reasons to make use of the Cell Signaling Technology western blotting protocol
NOTE: Please discuss with major antibody datasheet or product webpage for beneficial major antibody dilution buffer and beneficial antibody dilution.
A. Solutions and Reagents
From pattern preparation to detection, the reagents you want in your Western Blot are actually in a single handy package: #12957 Western Blotting Application Solutions Kit
Learn about our Solutions and Reagents
NOTE: Prepare options with reverse osmosis deionized (RODI) or equal grade water.
- 20X Phosphate Buffered Saline (PBS): (#9808) To put together 1 L 1X PBS: add 50 ml 20X PBS to 950 ml dH2O, combine.
- 10X Tris Buffered Saline (TBS): (#12498) To put together 1 L 1X TBS: add 100 ml 10X to 900 ml dH2O, combine.
- 1X SDS Sample Buffer: Blue Loading Pack (#7722) or Red Loading Pack (#7723) Prepare contemporary 3X lowering loading buffer by including 1/10 quantity 30X DTT to 1 quantity of 3X SDS loading buffer. Dilute to 1X with dH2O.
- 10X Tris-Glycine SDS Running Buffer: (#4050) To put together 1 L 1X operating buffer: add 100 ml 10X operating buffer to 900 ml dH2O, combine.
- 10X Tris-Glycine Transfer Buffer: (#12539) To put together 1 L 1X Transfer Buffer: add 100 ml 10X Transfer Buffer to 200 ml methanol + 700 ml dH2O, combine.
- 10X Tris Buffered Saline with Tween® 20 (TBST): (#9997) To put together 1 L 1X TBST: add 100 ml 10X TBST to 900 ml dH2O, combine.
- Nonfat Dry Milk: (#9999).
- Blocking Buffer: 1X TBST with 5% w/v nonfat dry milk; for 150 ml, add 7.5 g nonfat dry milk to 150 ml 1X TBST and blend nicely.
- Wash Buffer: (#9997) 1X TBST.
- Bovine Serum Albumin (BSA): (#9998).
- Primary Antibody Dilution Buffer: 1X TBST with 5% BSA or 5% nonfat dry milk as indicated on major antibody datasheet; for 20 ml, add 1.Zero g BSA or nonfat dry milk to 20 ml 1X TBST and blend nicely.
- Biotinylated Protein Ladder Detection Pack: (#7727).
- Prestained Protein Marker, Broad Range (11-190 kDa): (#13953).
- Blotting Membrane and Paper: (#12369) This protocol has been optimized for nitrocellulose membranes. Pore measurement 0.2 µm is mostly beneficial.
- Secondary Antibody Conjugated to HRP: anti-rabbit (#7074); anti-mouse (#7076).
- Detection Reagent: LumiGLO® chemiluminescent reagent and peroxide (#7003) or SignalFire™ ECL Reagent (#6883).
B. Protein Blotting
A normal protocol for pattern preparation.
Sample prep, SDS-PAGE and switch
- Treat cells by including contemporary media containing regulator for desired time.
- Aspirate media from cultures; wash cells with 1X PBS; aspirate.
- Lyse cells by including 1X SDS pattern buffer (100 µl per nicely of 6-well plate or 500 µl for a 10 cm diameter plate). Immediately scrape the cells off the plate and switch the extract to a microcentrifuge tube. Keep on ice.
- Sonicate for 10–15 sec to finish cell lysis and shear DNA (to scale back pattern viscosity).
- Heat a 20 µl pattern to 95–100°C for five min; cool on ice.
- Microcentrifuge for five min.
- Load 20 µl onto SDS-PAGE gel (10 cm x 10 cm).NOTE: Loading of prestained molecular weight markers (#13953, 5 µl/lane) to confirm electrotransfer and biotinylated protein ladder (#7727, 10 µl/lane) to find out molecular weights are beneficial.
- Electrotransfer to nitrocellulose membrane (#12369).
C. Membrane Blocking and Antibody Incubations
Block and Antibody Incubations
NOTE: Volumes are for 10 cm x 10 cm (100 cm2) of membrane; for various sized membranes, alter volumes accordingly.
I. Membrane Blocking
- (Optional) After switch, wash nitrocellulose membrane with 25 ml TBS for five min at room temperature.
- Incubate membrane in 25 ml of blocking buffer for 1 hr at room temperature.
- Wash thrice for five min every with 15 ml of TBST.
II. Primary Antibody Incubation
Proceed to one of many following particular set of steps relying on the first antibody used.
For Unconjugated Primary Antibodies
- Incubate membrane and first antibody (on the acceptable dilution and diluent as beneficial within the product datasheet) in 10 ml major antibody dilution buffer with mild agitation in a single day at 4°C.
- Wash thrice for five min every with 15 ml of TBST.
- Incubate membrane with the species acceptable HRP-conjugated secondary antibody (#7074 or #7076 at 1:2000) and anti-biotin, HRP-linked Antibody (#7075 at 1:1000–1:3000) to detect biotinylated protein markers in 10 ml of blocking buffer with mild agitation for 1 hr at room temperature.
- Wash thrice for five min every with 15 ml of TBST.
- Proceed with detection (Section D).
For HRP Conjugated Primary Antibodies
- Incubate membrane and first antibody (on the acceptable dilution as beneficial within the product datasheet) in 10 ml major antibody dilution buffer with mild agitation in a single day at 4°C.
- Wash thrice for five min every with 15 ml of TBST.
- Incubate with Anti-biotin, HRP-linked Antibody (#7075 at 1:1000–1:3000), to detect biotinylated protein markers, in 10 ml of blocking buffer with mild agitation for 1 hr at room temperature.
- Wash thrice for five min every with 15 ml of TBST.
- Proceed with detection (Section D).
For Biotinylated Primary Antibodies
- Incubate membrane and first antibody (on the acceptable dilution as beneficial within the product datasheet) in 10 ml major antibody dilution buffer with mild agitation in a single day at 4°C.
- Wash thrice for five min every with 15 ml of TBST.
- Incubate membrane with Streptavidin-HRP (#3999 on the acceptable dilution) in 10 ml of blocking buffer with mild agitation for 1 hr at room temperature.
- Wash thrice for five min every with 15 ml of TBST.
- Proceed with detection (Section D).
Do not add Anti-biotin, HRP-linked Antibody for detection of biotinylated protein markers. There is not any want. The Streptavidin-HRP will even visualize the biotinylated markers.
D. Detection of Proteins
Protein Detection
- Incubate membrane with 10 ml LumiGLO® (0.5 ml 20X LumiGLO® #7003, 0.5 ml 20X Peroxide, and 9.Zero ml purified water) or 10 ml SignalFire™ #6883 (5 ml Reagent A, 5 ml Reagent B) with mild agitation for 1 min at room temperature.
- Drain membrane of extra growing resolution (don’t let dry), wrap in plastic wrap and expose to x-ray movie. An preliminary 10 sec publicity ought to point out the right publicity time.NOTE: Due to the kinetics of the detection response, sign is most intense instantly following incubation and declines over the next 2 hr.